Overview
EpiBibR stands for “epidemiology-based bibliography for R.” The R package is under the MIT License and as such is a free resource based on the open science principles (reproducible research, open data, open code). The resource may be used by researchers, whose domain is scientometrics, but also by researchers from other disciplines. For instance, the scientific community in Artificial Intelligence and Data Science may use this package to accelerate new research insights about covid-19. The package follows the methodology put in place by the Allen Institute and its partners to create the CORD-19 dataset with some differences. The later is accessible through downloads of sub-sets or through a REST API. The data provide important information such as authors, methods, data, and citations to make it easier for researchers to find relevant contributions to their research questions. Our package proposes 22 features for the 145,415 references (on July 6, 2021) and access to the data has been made as easy as possible in order to integrate efficiently in almost any researcher’s pipeline.
Through this package, a researcher can connect the data to her analysis based on the R language. With this workflow in mind, a researcher can save time on collecting data, and can use a very accessible language to perform complex analytical tasks, be it R or Python for instance. Indeed, it is usual that researchers use multiple languages (functional or not) to produce certain outputs. This workflow opens these data to analyses from the largest spectrum of potential options, enhancing multidisciplinary approaches applied to these data (biostatistics, bibliometrics, text mining, etc.).
The goal of this package in this emergency context is to limit the references to the medical domain, hence limiting ourselves to the Pubmed repository, but then to leverage the methodologies used across different disciplines. As we will address this point later, a further extension could be to add references from other disciplines to not only benefit from the wealth of methodologies but also from their own theories and concepts. For instance, to assess the spread of the disease, the literature - and theories - from researchers in demography would certainly be relevant.
Functionality
The references were collected via PubMed, a free resource that is developed and maintained by the National Center for Biotechnology Information (NCBI), at the U.S. National Library of Medicine (NLM), located at the National Institutes of Health (NIH). PubMed includes over 30 million citations from biomedical literature.
More specifically, to collect our references, we adopted the procedure used by the Allen Institute for AI for their CORD-19 project. We apply a similar query on PubMed: “COVID-19” OR Coronavirus OR “Corona virus” OR “2019-nCoV” OR “SARS-CoV” OR “MERS-CoV” OR “Severe Acute Respiratory Syndrome” OR “Middle East Respiratory Syndrome” to build our own bibliographic data.
To navigate through our dataset, EpiBibR relies on a set of search arguments: author, author’s country of origin, keyword in the title, keyword in the abstract, year and the name of the journal. Each of them can truly help scientists and R users to filter references and find the relevant articles.
In an effort to simplify the workflow between our package and the research methodologies, the format of our dataframe has been designed to integrate with different data pipelines, notably to facilitate the use of the R package Bibliometrix with our data (Aria and Cuccurullo 2017).
Features
| Field Tags | Descriptions | Field Tags | Descriptions |
|---|---|---|---|
| AU | Authors | ISSN | Source Code |
| TI | Document Title | VOL | Volume |
| AB | Abstract | ISSUE | Issue Number |
| PY | Year | LT | Language |
| DT | Document Type | C1 | Author Address |
| MESH | Medical Subject Headings Vocabulary | RP | Reprint Address |
| TC | Times Cited | ID | PubMed ID |
| SO | Publication Name (or Source) | DE | Authors’ Keywords |
| J9 | Source Abbreviation | UT | Unique Article Identifier |
| JI | ISO Source Abbreviation | AU_CO | Author’s Country of Affiliation |
| DI | Digital Object Identifier (DOI) | DB | Bibliographic Database |
Practical usage
Quick start
First, install EpiBibR:
devtools::install_github("warint/EpiBibR")Next, call EpiBibR to make sure everything is installed correctly.
How-To
EpiBibR allows you to search bibligraphic references using several arguments : Author, author’s country of origin, author + year, keywords in the title, keywords in the abstract, year and source name.
Retrieve bibliographic data
To get the entire bibliographic dataframe contaning more than 80 000 references, use the epibib_data function.
complete_data <- epibibr_data()Search by keywords in title
covid_articles <- epibibr_data(title = "covid")As you may have noticed, you can keep more than one argument to refine your search.
Search by keywords in title and by year
covid2020_articles <- epibibr_data(title = "covid", year = "2020")Search by keywords in the abstract
coronavirus_articles <- epibibr_data(abstract = "coronavirus")Search by year
A2020_articles <- epibibr_data(year = "2020")Search by source
bio_articles <- epibibr_data(source = "bio")Create visuals
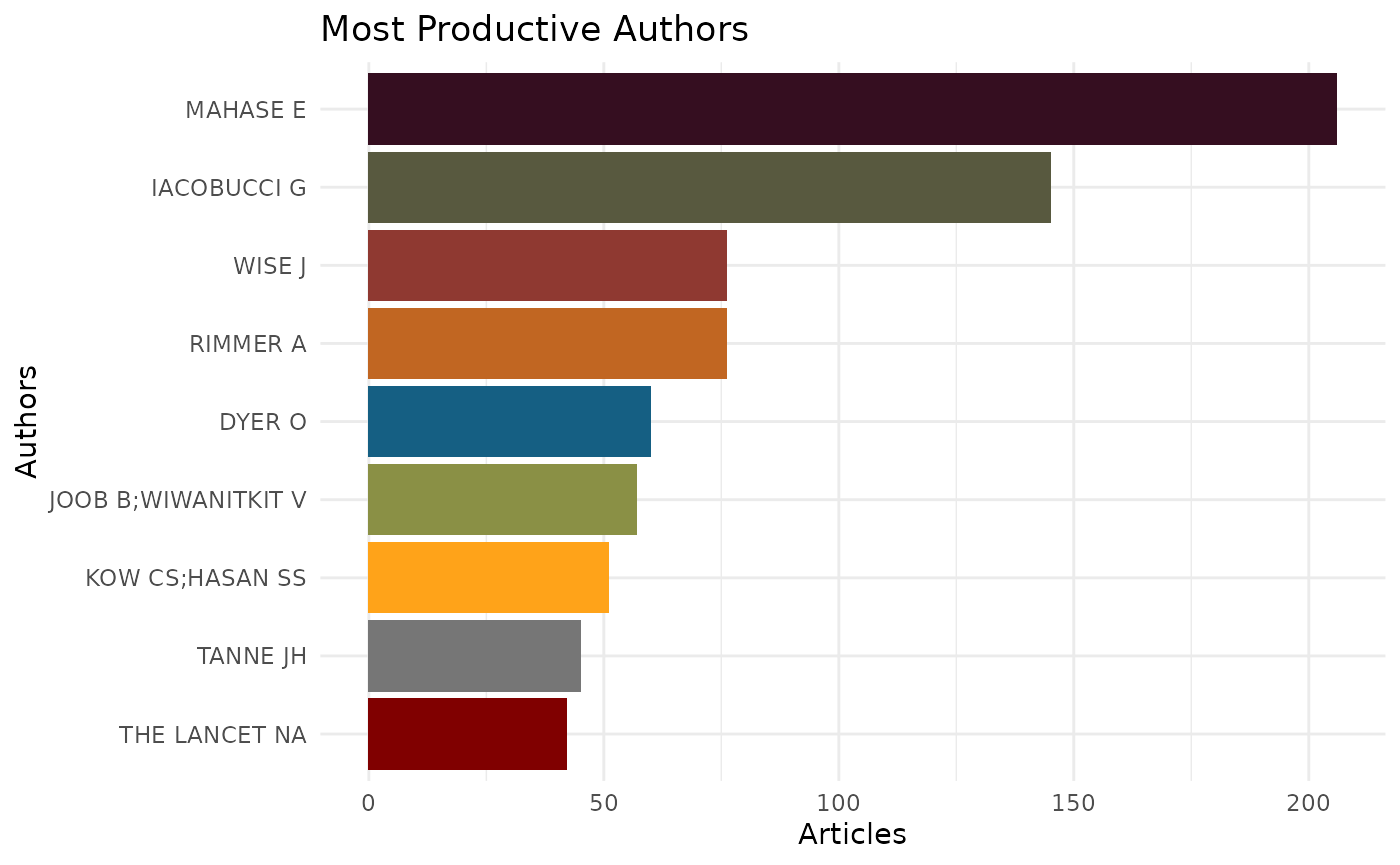
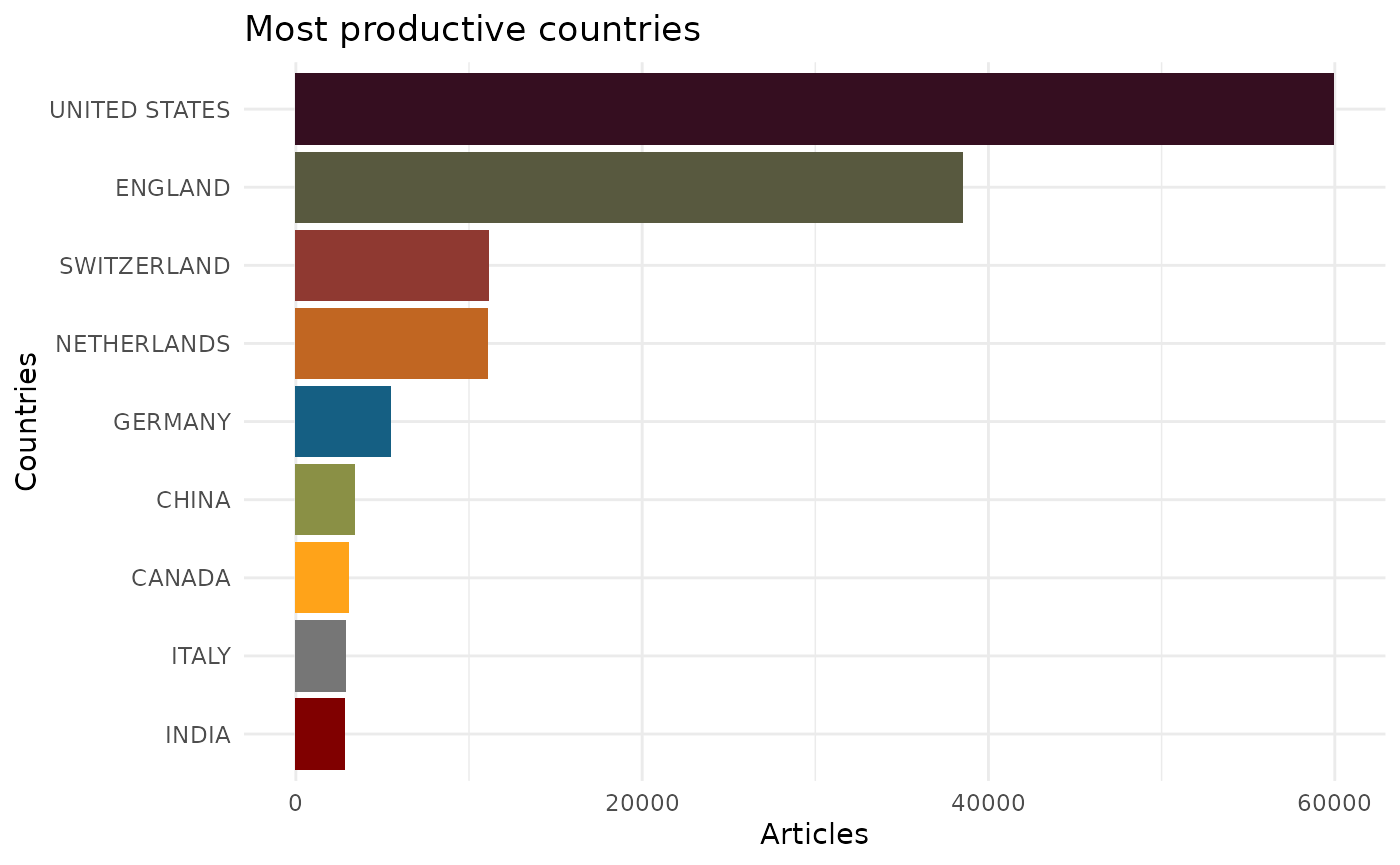
EpiBibR allows you to create three summary visuals on its data.
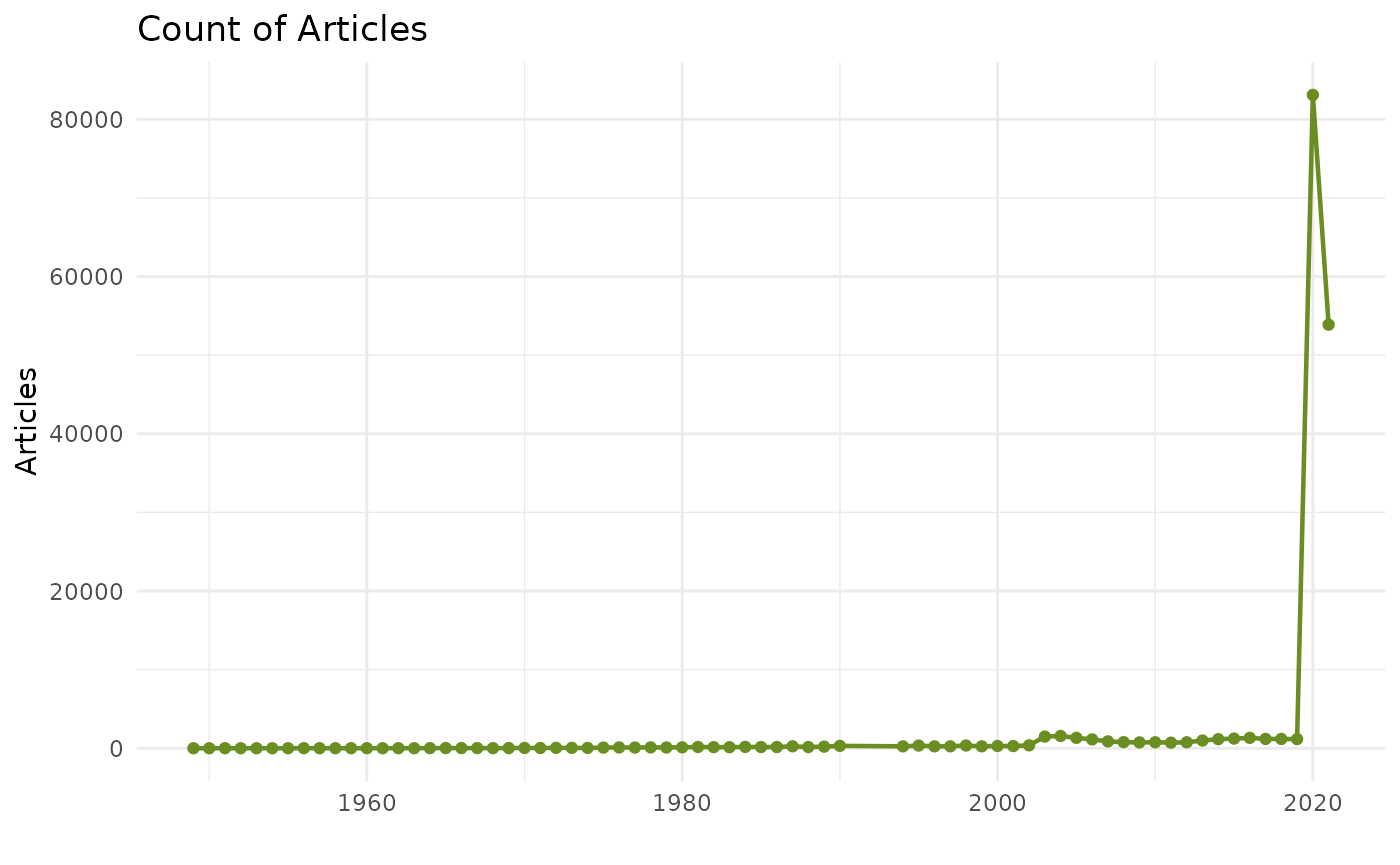
Number of articles
**Note that if the argument title is set to TRUE or is empty, title will appear. To hide it, title must be set to FALSE
epibibr_visual(chart = "line_1", title = TRUE)